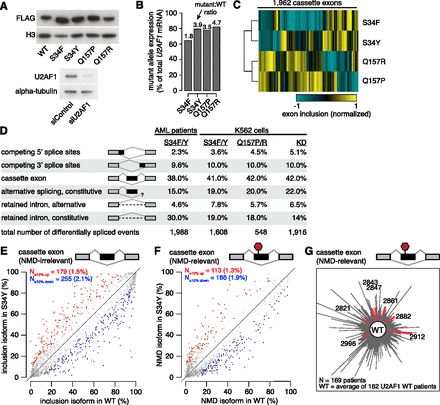
U2AF1 mutations alter splicing, but do not cause splicing failure. (A) Western blots showing levels of FLAG-tagged U2AF1 in K562 cells stably expressing the indicated alleles (top) and levels of endogenous U2AF1 in K562 cells following transfection with a nontargeting siRNA or a siRNA pool against U2AF1 (bottom). (B) U2AF1 mutant allele expression as a percentage of total U2AF1 mRNA in K562 cells. (C) Heat map of K562 cells expressing mutant U2AF1. Dendrogram is from an unsupervised cluster analysis based on cassette exon inclusion levels. (D) U2AF1 mutation-dependent changes in splicing for AML S34 versus WT patients, K562 S34F or S34Y versus WT expression, K562 Q157P or Q157R versus WT expression, and K562 U2AF1 KD versus control KD. Percentages indicate the fraction of mutation-dependent splicing changes falling into each category of splicing event. (E) Levels of cassette exon inclusion in K562 cells expressing WT or S34Y U2AF1. (N) Numbers of alternatively spliced cassette exons with increased/decreased inclusion; (percentages) fraction of alternatively spliced cassette exons that are affected by S34Y expression. Events that do not change significantly are rendered transparent. Plot restricted to cassette exon events that are predicted to not induce nonsense-mediated decay (NMD). (F) Levels of NMD-inducing isoforms of cassette exon events in K562 cells expressing WT or S34Y U2AF1. (G) Levels of NMD-inducing isoforms of cassette exon events in AML transcriptomes. Distance from the center measures the splicing dissimilarity between each AML transcriptome and the average of all U2AF1 WT samples, defined as the sum of absolute differences in expression of NMD-inducing isoforms.











