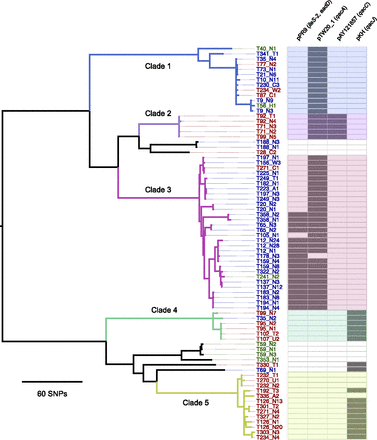
Existence of circulating clades of MRSA ST 239. Maximum likelihood phylogenetic tree based on core genome SNPs of the first and up to two repeat isolates from 46 patients and five healthcare workers. The first T number assigned to each represents the case study number. The second letter/number combination refers to the type of sample and the sample number from that particular site: (N) nasal swab; (T) throat swab; (A) axillae swab; (W) wound swab; (U) urine; (C) tracheal suction. For example, T178-N3 refers to the third nasal swab taken from case T178. Isolate labels are color-coded to signify isolation from a staff member (green), a patient on the adult ICU (red), or pediatric ICU (blue). Also shown (right-hand panel) is the presence (bold) or absence (pale) of four plasmids harboring antibiotic and antiseptic resistance genes. The colors used to denote clades 1 to 5 are cross-referenced in Figure 3.











