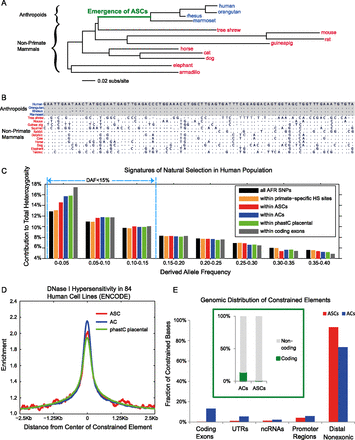
Functionality and genomic distribution of ASCs. (A) Phylogenetic tree: anthropoids (blue) and nonprimate mammals (red). (B) Illustrative example: Sequence alignment of ASC19060 shows constraint in anthropoids and unconstrained divergence among nonprimate mammals. Dots represent identical nucleotides. (C) Derived allele frequency spectra of African (AFR) SNPs from the 1000 Genomes Project. SNPs within ASCs are shifted to lower frequencies (<15%), indicating ongoing negative selection in humans. In contrast, SNPs within biochemically defined primate-specific DNase I hypersensitive (HS) sites (Jacques et al. 2013) show only a weak frequency shift. (D) ASCs are enriched for DNase I hypersensitivity in 84 human cell lines. (E) Distribution of constrained elements in the human genome. (Green subplot) Of ASC base pairs, 0.3% are protein-coding relative to 13.1% for ACs.











