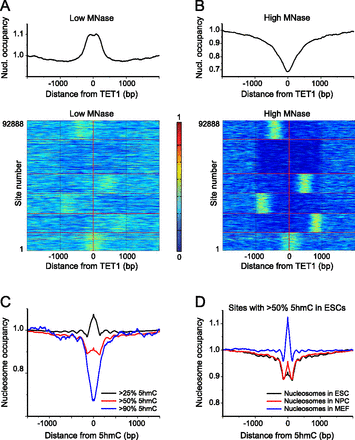
Nucleosome occupancy around TET1 binding sites and in relation to 5hmC levels. (A) Average nucleosome occupancy (top panel) and k-means cluster plots showing nucleosome occupancy around each of 92,888 TET1 ChIP-seq peaks in ESCs (Yu et al. 2012) at low MNase digestion (bottom panel). (B) Same as panel A but for high MNase digestion. (C) Aggregate plot of nucleosome occupancy around 5hmC sites in ESCs (Yu et al. 2012) grouped according to their 5hmC levels as >25% 5hmC (black line), >50% 5hmC (red line), and > 90% 5hmC (blue line). Upon increasing the 5hmC level, the nucleosome density changed from slight enrichment to nucleosome depletion, which corresponds to the nucleosome removal at a subset of these sites. (D) Changes of the nucleosome occupancy during cell development (ESCs, NPCs, and MEFs) around hydroxymethylated sites in ESCs (>50% 5hmC).











