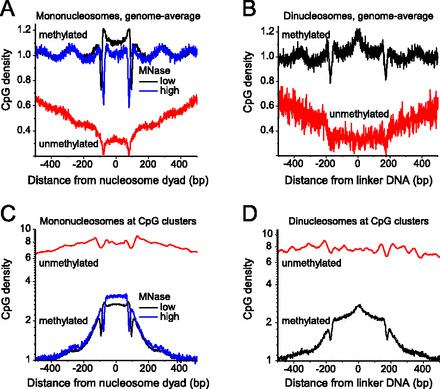
DNA methylation patterns relative to nucleosomes. (A) Average CpG density of methylated (>50% 5mC) and unmethylated (<10% 5mC) mononucleosome sequences in ESCs for the complete genome. Most of the sequences reside outside of CGIs because the latter represent only a fraction of ∼1% of the total pool of sequences mapped by MNase-seq. (Black line) DNA methylation along the nucleosomes obtained at low MNase digestion; (blue line) high MNase digestion. The nucleosome dyad is the middle of the nucleosomal DNA fragment. (B) Same as in panel A but for the dinucleosome fraction. The linker is defined as the middle of the dinucleosome. (C) CpG density for mononucleosomes and associated linker DNA inside CGIs. Putative CGIs were taken from the coordinates of 125,303 CpG clusters defined by the proximity of neighboring CpGs with the CpGcluster2 algorithm (Hackenberg et al. 2006). (D) Same as in panel C but for the dinucleosome fraction.











