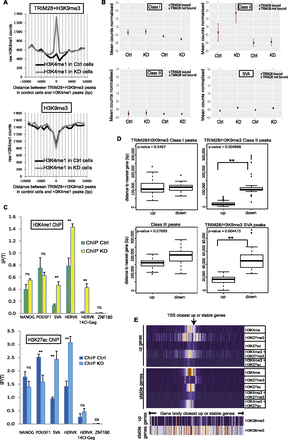
TRIM28 prevents enhancer activation marks at EREs and modulates the local transcriptional landscape in hESC. (A) ChIP-Cor analysis demonstrating that H3K9me3 at TRIM28-bearing loci (top panel) is replaced by H3K4me1 upon TRIM28 KD, while this does not occur at TRIM28-independent H3K9me3-positive sites (bottom panel). (B) H3K4me1 ChIP-seq analysis on sh-NS control (Ctrl) and TRIM28 KD cells, giving counts associated with TRIM28-bound or -devoid EREs. The total number of reads per 106 bases of ERE is normalized to the total number of reads in the ChIP-seq and to the number of reads found in the respective inputs. TRIM28-bound Class II and SVA elements significantly gain H3K4me1 after TRIM28 KD (Wilcoxon test gave P-values < 0.0001 for Class II and SVA but no significance for Class I and Class III HERVs). (C) H3K4me1 (top panel) and H3K27ac (bottom panel) ChIP-qPCR on control and TRIM28 KD cells using either generic primers detecting SVA and HERVK families or primers detecting HERVK14CI members. Primers targeting the 3′ end of ZNF180 and those targeting NANOG and POU5F1 promoters are used as negative and positive controls, respectively. Values were normalized to their respective total inputs (IP/Ti) and to the positive control GAPDH. Bars represent the mean and SD of technical replicates (n = 3). T-tests were used to compare controls and TRIM28-depleted samples. ([**] P-value ≤ 0.01). (D) Average distance between genes up- or down-regulated upon TRIM28 depletion and indicated genomic features. Up-regulated genes had a statistically higher chance of being close to a TRIM28/H3K9me3 bearing Class II HERVs or SVAs. Wilcoxon Mann-Whitney nonparametric test P-values are indicated. (E) Indicated histone marks ChIP-seq data from the ENCODE Project found within 10 kb (in 100-bp bins; orange and purple correspond to high and low relative occupancy, respectively) of the TSS (top) or over the transcribed region (bottom) of TRIM28-bound, H3K9me3-bearing, SVA-close genes, either up-regulated (up) or unaffected (stable) upon TRIM28 KD.











