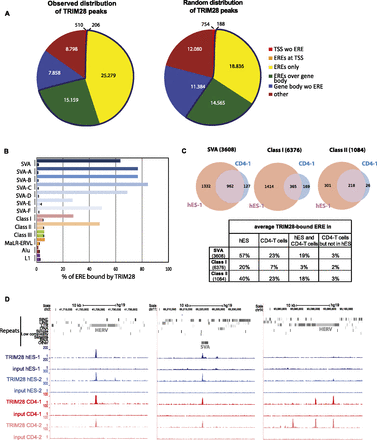
TRIM28 is associated with Class I and II HERVs and SVAs in hESC. (A) Observed and expected distribution of TRIM28 ChIP-seq peaks among indicated entities as annotated on the UCSC Genome Browser. Only HERVs containing both LTR and internal sequences were included in the analysis and counted as one full-length single element (see repeats analysis in Methods for details). Number of peaks for each category is indicated. The blue line delineates the fraction with ERE elements. (TSS) Transcription start site ±500 bp; (ERE) endogenous retroelement. (B) Relative TRIM28 occupancy of different ERE families. For each ERE (as defined in A), both the values observed (top bar) and expected by chance (bottom bar, as obtained from averaging a 10-times randomization of TRIM28 locations) are shown. For SVAs, data for both the whole family and its different subgroups are given. The abundant and heterogeneous MaLR-ERVL elements were monitored separately from the HERV class III. ChIP-seq data from an independent biological replicate are illustrated in Supplemental Figure S1A. (C) Comparison of TRIM28 binding to indicated EREs in human ES cells and CD4+ T-lymphocytes. In parentheses, numbers of EREs as defined in A are given. The ChIP was performed on T-cells pooled from three independent donors. The Venn diagrams represent data generated from one hES and one CD4+ T-cell ChIP-seq (hES-1 and CD4-1). Similar results were obtained from hESC and T-cell biological replicates (hES-2 and CD4-2) (Supplemental Fig. S1B). Diagrams obtained from replicates are illustrated in Supplemental Figure S1B. The table gives the average of the values obtained with all the replicates. (D) ChIP-seq maps of TRIM28 providing examples of EREs bound both in ES and CD4+ T-cells or in CD4+ T-cells only. Two biological replicates are illustrated in each case (hES-1, hES-2 and CD4-1, CD4-2) and displayed on the UCSC Genome Browser with the RepeatMasker function disabled to visualize EREs. Control maps obtained for the sequencing of the respective inputs are shown (set to the same vertical scale).











