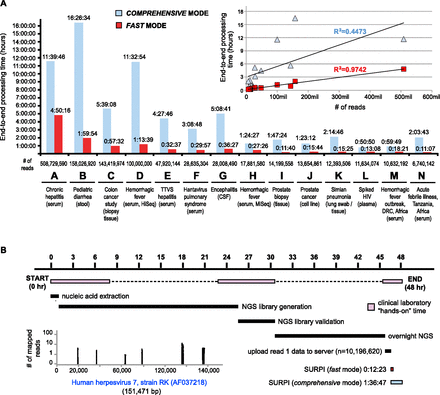
Speed of SURPI and feasibility for real-time clinical analysis. (A) Timing performance for SURPI in fast mode (red) and comprehensive mode (blue) was benchmarked on a single computational server across 12 NGS data sets representing a variety of infectious diseases and sample types. Processing end-to-end-times are plotted against the number of reads (inset), along with regression trend lines corresponding to SURPI processing in fast and comprehensive modes. (B) A serum sample from a returning traveler with an acute febrile illness was analyzed using NGS, resulting in SURPI detection of human herpesvirus 7 (HHV-7) infection (inset, coverage plot) in a clinically relevant 48-h timeframe.











