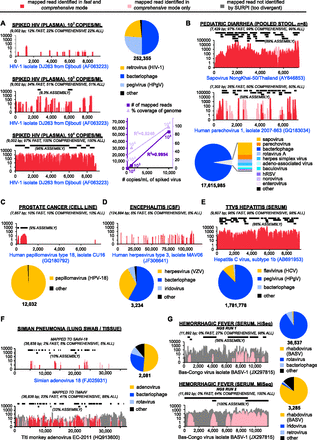
The SURPI pipeline correctly identifies viral species in clinical NGS data sets. Data sets corresponding to clinical samples or sample pools harboring target viral pathogens were analyzed using SURPI. Pie charts show detected viruses derived from the output summary tables. Target viruses are color-coded in yellow or orange; other viruses are color-coded ranked by their relative abundance in shades of blue, followed by shades of purple. Coverage maps of the “best hit” viral genome in fast mode (red) and comprehensive mode (pink, overlaid by red) display automated SURPI output corresponding to the detected target viral genome (blue text). The read coverage (y-axis, log scale) and de novo assembled contigs (black lines) are plotted as a function of nucleotide position along the genome (x-axis). Percent coverage achieved using SURPI in fast mode (“FAST”), in comprehensive mode (“COMPREHENSIVE”), and by de novo assembly (“ASSEMBLY”), as well as the actual coverage from all reads in the data set (“ALL”) are shown. (A) Coverage plots of HIV-1 spiked at titers of 102−104 copies/mL. The number of mapped reads and percent coverage are plotted against the viral copy number (inset). Coverage plots of SaV and HPeV-1 (B), HPV-18 (C), HHV-3 (D), and HCV-1b (E). (F) Coverage plot mapping SURPI-classified genus-level Mastadenovirus reads (red/pink) to the SAdV-18 genome, or Mastadenovirus reads (red/pink) and all specific TMAdV reads (gray) to the TMAdV genome. (G) Coverage plots mapping SURPI-classified family-level Rhabdoviridae reads (pink) or all specific BASV reads (gray) to the BASV genome.











