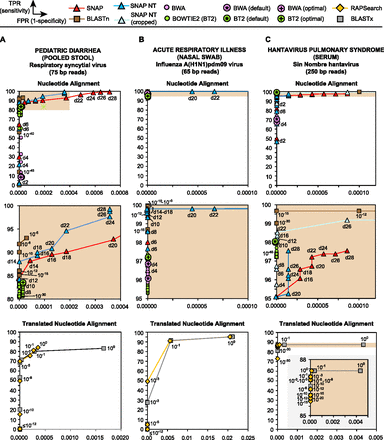
SURPI aligners (SNAP and RAPSearch) are comparable to other tested aligners for detection of viral reads in clinical NGS data sets. ROC curves were generated to evaluate the ability of nucleotide and translated nucleotide (protein) aligners to detect reads corresponding to three target viruses: (A) respiratory syncytial virus (RSV) from stool; (B) influenza A(H1N1)pdm09 from a nasal swab; and (C) Sin Nombre hantavirus from serum. Sensitivity or the true positive rate (TPR) (y-axis) is plotted against 1-specificity or the false positive rate (FPR) (x-axis). For each aligner, reads assigned to the correct viral genus were used for generating the ROC curve. The shaded panels are magnifications of the corresponding shaded regions in the upper panels (A–C, nucleotide alignment) or overlapping larger panel (C, translated nucleotide alignment).











