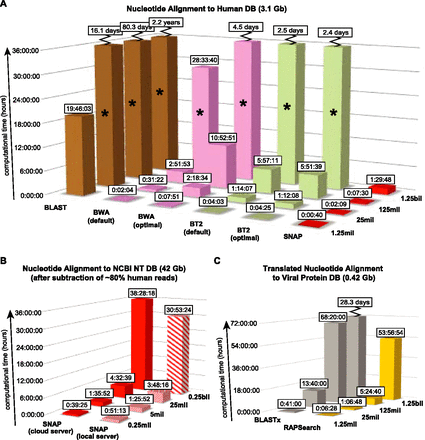
SURPI aligners (SNAP and RAPSearch) are significantly faster than other tested aligners and scale better with larger data sets. Timing performance was benchmarked on a single computational server using in silico query data sets of increasing size. The breaks (zigzag lines) represent computational times that are off-scale. Some of the computational times were estimated (asterisks). (A) Performance time for alignment of reads to the human DB. (B) Performance time for SNAP alignment of reads to the entire 42-Gb NCBI nt DB. The z-axis denotes the approximate number of remaining reads following computational subtraction against the human DB. SNAP performance times were benchmarked separately on local and cloud servers. (C) Performance times for translated nucleotide alignment to the viral protein DB using RAPSearch and BLASTx.











