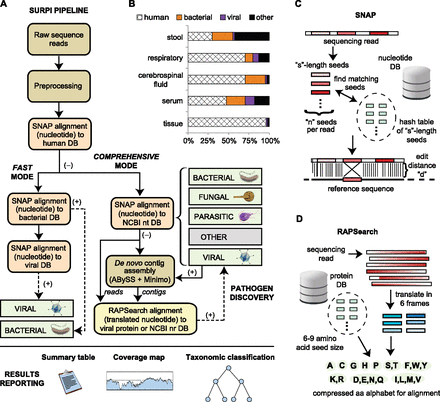
The SURPI pipeline for pathogen detection. (A) A schematic overview of the SURPI pipeline. Raw NGS reads are preprocessed by removal of adapter, low-quality, and low-complexity sequences, followed by computational subtraction of human reads using SNAP. In fast mode, viruses and bacteria are identified by SNAP alignment to viral and bacterial nucleotide databases. In comprehensive mode, reads are aligned using SNAP to all nucleotide sequences in the NCBI nt collection, enabling identification of bacteria, fungi, parasites, and viruses. For pathogen discovery of divergent microorganisms, unmatched reads and contigs generated from de novo assembly are then aligned to a viral protein database or all protein sequences in the NCBI nr collection using RAPSearch. SURPI reports include a list of all classified reads with taxonomic assignments, a summary table of read counts, and both viral and bacterial genomic coverage maps. (B) Relative proportion of NGS reads classified as human, bacterial, viral, or other in different clinical sample types. (C) The SNAP nucleotide aligner (Zaharia et al. 2011). SNAP aligns reads by generating a hash table of sequences of length “s” from the reference database and then comparing the hash index with “n” seeds of length “s” generated from the query sequence, producing a match based on the edit distance “d.” (D) The RAPSearch protein similarity search tool (Zhao et al. 2012). RAPSearch aligns translated nucleotide queries to a protein database using a compressed amino acid alphabet at the level of chemical similarity for greatly increased processing speed.











