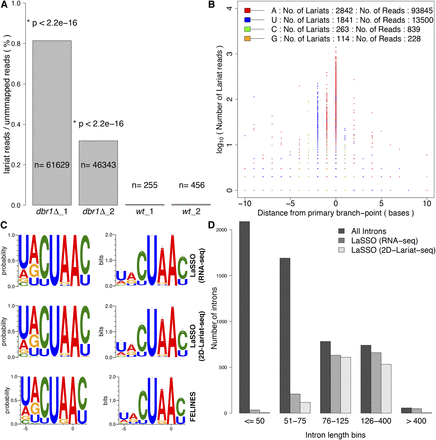
Characterization of lariat branch points and branch-site sequence. (A) Proportion of lariat reads relative to total number of reads not mapped to genome or transcriptome. Absolute numbers in each sample are indicated, along with P-values (Fisher’s exact test). (B) The base (color-coded as indicated) and position (x-axis) of each branch point identified as a function of read number supporting it (y-axis). The primary branch point is placed at position zero on the x-axis. The numbers of lariats and supporting reads are indicated on top. Only branch points located within 10 bases up- (negative values) or downstream (positive values) from a primary branch point are shown. (C) Consensus branch-site sequences around the primary branch point as probability (left) and bits (right), plotted using WebLogo (Crooks et al. 2004) (default settings except for compositional adjustment, with GC content set to 30%). (Top panels) Using LaSSO based on 1236 introns from our data; (middle panels) using LaSSO based on 930 introns from 2D-Lariat-seq data (Awan et al. 2013) that were supported by ≥3 lariat reads; (bottom panels) using FELINES for the same set of 1236 introns detected in this study. (D) Number of introns of different sizes for which lariat reads were detected by LaSSO when no read-number threshold was applied: 1584 lariats for our data, 1268 lariats for 2D-Lariat-seq data by Awan et al. (2013). Introns were binned according to their size as indicated (5361 introns in total).











