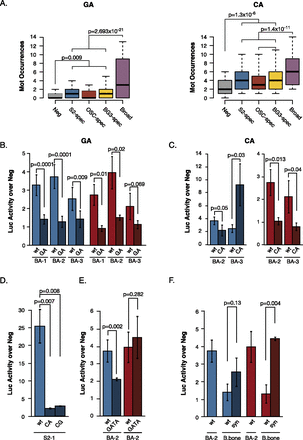
Dinucleotide repeat motifs are enhancer features required for activity. (A) Distribution of dinucleotide repeat motif (DRM) occurrences for the GA DRM Trl/ME137 (left) and the CA DRM Mot15 (right) in negative regions (gray), cell type-specific enhancers (blue, red, yellow), and broadly active enhancers (purple). The whiskers denote the 10th and 90th percentiles and Wilcoxon P-values are shown. (B–C) Luciferase (Luc) assays in S2 cells (blue) and OSCs (red) for three broadly active enhancers and DRM mutant variants. Shown are wild-type (light colors) and mutant (dark colors) sequences, in which GA DRMs (B) or CA DRMs (C) are disrupted. (D) Luciferase assays for disrupting the CA and CG DRMs in the cell type-specific enhancer S2-1. (E) Luciferase assays for disrupting the GATA motif in two broadly active enhancers. (B–E) Neg denotes a negative control sequence used for normalization (Arnold et al. 2013), and error bars indicate standard deviations of at least three independent biological replicates. Shown are P-values from unpaired t-tests. (F) Luciferase assays in S2 cells (blue) and OSCs (red) of a synthetic enhancer (syn) for which the GA and CA DRMs and the AP-1 motif were copied from a Broad enhancer (BA-2) into an inactive genomic region (B.bone, backbone), while preserving their orientation and spacing. The activities of BA-2 and B.bone are shown as controls.











