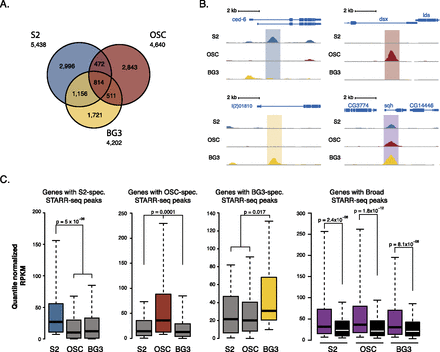
STARR-seq identifies enhancers with different cell type activity profiles. (A) Venn diagram of the STARR-seq enhancers according to their activity in three different Drosophila cell types: S2 (blue), OSC (red), and BG3 (yellow). (B) UCSC Genome Browser screenshot for examples of S2-specific, OSC-specific, BG3-specific, and broadly active enhancers. (C) The expression levels of genes neighboring enhancers from each of the four enhancer classes (quantile-normalized RPKM [reads per kilobase exon model] values; Wilcoxon P-values) in S2 cells, OSCs, and BG3 cells. Black shows a negative control with genes neighboring randomly chosen regions that were inactive in all three cell types, according to STARR-seq.











