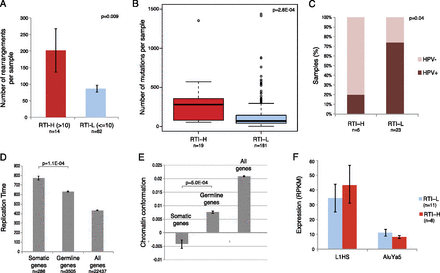
Retrotransposon load is correlated with genomic instability, late-replication, and closed chromatin. (A) Number of somatic rearrangements in LUSC, LUAD, and HNSC samples with high retrotransposon load (>10 somatic retrotransposon insertions, RTI-H) and with low retrotransposon load (≤10 somatic insertion, RTI-L). (B) Number of somatic mutations in RTI-H and RTI-L samples across all 11 tumor types. (C) HPV status of RTI-H and RTI-L HNSC samples. (D) Replication timing of genes that contain somatic retrotransposon insertions versus genes that contain germline insertions, and all RefSeq genes. Later replicating genes have higher values of replication time on the y-axis. (E) Chromatin conformation of genes that contain somatic retrotransposon insertions versus genes that contain germline insertions, and all RefSeq genes. The y-scale represents relative chromatin “openness,” the lower the y-value, the more closed the chromatin state. (F) Expression (RPKM) of consensus L1HS and AluYa5 sequences in RTI-H and RTI-L LUSC samples. All error bars represent standard error of the distribution.











