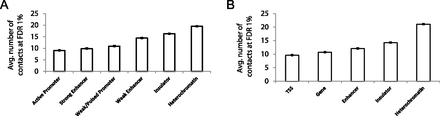
Insulators and heterochromatin regions participate in more high-confidence contacts than promoters and strong enhancers. Average number of high-confidence (FDR 1%) contacts and their standard errors identified by Fit-Hi-C for each annotation term from two different semiautomated genome annotation methods. Contact confidences are assigned at a resolution of 10 RE fragments for the Hi-C data from hESC cells (Dixon et al. 2012), using ICE-corrected contact maps and one-step refinement of the null model (spline-2). In order to map genome annotations to the windows used in the Fit-Hi-C analysis, each annotated region is assigned to the 10 RE fragment window with which it has the most overlap. (A) Six selected annotations from 15-label genomic segmentation of hESC using ChromHMM (Ernst and Kellis 2012). (B) Five selected annotations from 25-label genomic segmentation of hESC using Segway (Hoffman et al. 2012, 2013). See Supplemental Figure 9 for plots with complete lists of labels for both segmentation methods.











