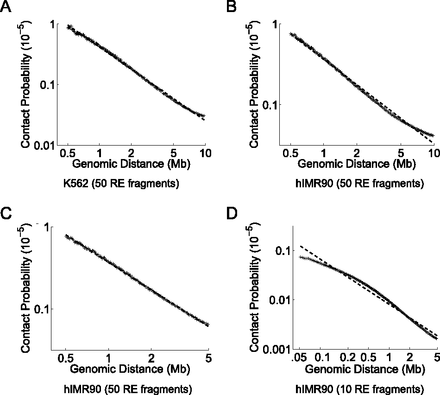
Differences in scaling of contact probability when sequencing depth, resolution, or the distance range of interest changes. We plot the relationship between contact probability and genomic distance by binning the total number of mid-range read pairs into 200 equal occupancy bins. We compute the mean genomic distance and mean contact probability among all locus pairs for each bin and calculate the best power-law (i.e., log-linear) fit to these observations (dashed line) for the genomic distance range spanned by the x-axis. We plot these for the K562 library from Lieberman-Aiden et al. (2009) at a resolution of 50 RE fragments (A), for the hIMR90 library from Dixon et al. (2012) at a resolution of 50 RE fragments for genomic distances up to 10 Mb (B), 50 RE fragments for genomic distances up to 5 Mb (C), and 10 RE fragments (D).











