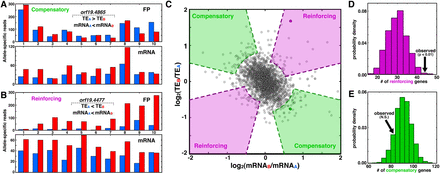
Biases in transcription and translation are often coordinated, with interactions favoring compensation over reinforcing. (A,B) Specific examples of compensatory (A) and reinforcing (B) interactions between transcription and translation. Genes with compensatory interactions have higher allelic difference at the mRNA level than at the FP level, whereas those with reinforcing interactions differ more at the FP level than at the mRNA level. (C) Scatterplot of mRNAALFD and TEALFD levels, where genes from A and B are indicated as darkened circles, and shaded regions indicate compensatory and reinforcing interactions, as indicated. The purple and green regions’ curved portions reflect the two-standard-deviation spread of the data along the y = x and y = −x lines, respectively, and straight segments are based on a heuristic chosen to ensure that both the mRNAALFD and TEALFD values are nonzero (see Methods). (D,E) PDFs indicating the number of reinforcing (D) and compensatory (E) genes from permuted data; arrows indicate the number of genes from the observed data.











