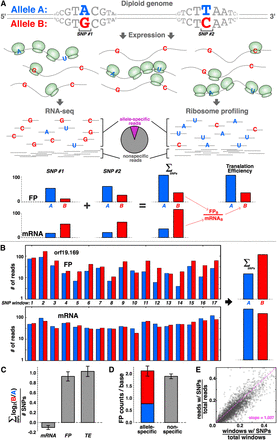
Sensitively detecting ASE at translational level with ribosome profiling. (A) Schematic of the approach. For a given gene with two SNPs, transcripts from the B allele may be more abundant, whereas translation favors the A allele, as indicated by increased density of ribosomes, shown in green. These biases are revealed by RNA-seq and ribosome profiling, respectively. Allele-specific reads are summed across all SNPs in the gene, and translational efficiency (“TE”) is calculated from the mRNA and footprint (“FP”) levels. (B,C) Signal is consistent across many SNPs. There are 17 distinct SNP windows in orf19.169/CHO2, and the majority indicates a translational bias toward the B allele, but roughly equal transcript levels (B), with little error across SNPs (C); error bars, ±SEM. (D) The sum of allele-specific reads (red and blue bar) matches the level of nonspecific reads that do not include SNPs (gray bar) for orf19.169; error bars, ±SEM. (E) Across all genes, the fraction of SNP-containing reads corresponds strongly to the fraction of gene length comprised of SNP-containing windows.











