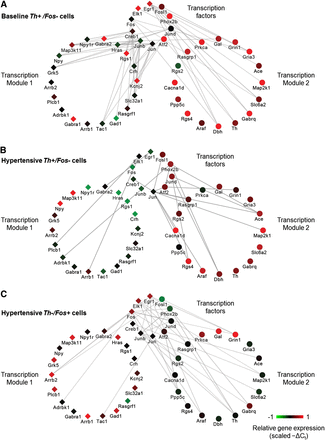
Gene correlation networks. The correlative network structures represent correlative relationships shared between TFs and target genes of each module across the three cell types: baseline Th+, hypertension Th+, hypertension Fos+. Cytoscape software was used to visualize the correlative network relationships. Edge opacity represents the strength of the correlation shared between genes across the respective sample subset (e.g., Th+/Fos− single cells): the darker the edge, the higher the correlation coefficient values. These network structures illustrate the pairwise Spearman rank correlative relationships among the subset of 48 genes. TFs are separated from the subset while the remaining genes are organized into their respective transcription modules 1 and 2 (Fig. 4C). The correlation network is based on pairwise gene correlations across various subsets of single cells. Only pairwise Spearman correlation coefficients ≥0.4 were included. Node colors represent scaled −ΔCt values of a representative single cell sample from the respective neuronal subtype. (A) Pairwise gene correlation network across normotensive single cells. Note the high number of correlative relationships shared between TFs and genes from both modules 1 and 2. (B) Correlation network based on hypertensive Th+/Fos− single cells shows a significant change in the number of correlative relationships between TFs and downstream target genes, and the majority of these relationships exist between TFs and genes within module 2. Similarly, this same shift in pairwise relationships occurs in Th−/Fos+ single cells, shown in C. This shift in relationships suggests that a physiological perturbation, in this case acute hypertension, causes a shift in the correlative relationships between TFs and downstream genes.











