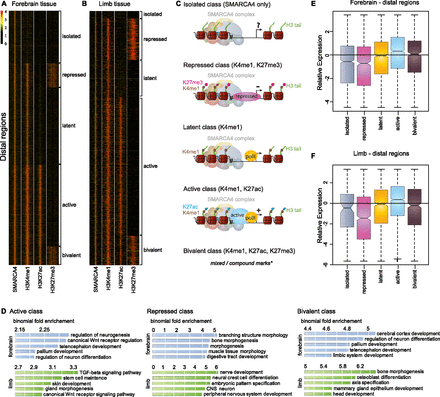
Histone marks associated with activation and repression co-localize with SMARCA4. (A,B) Heatmaps displaying coverage for SMARCA4 and informative histone marks for proximal (H3K4me3 and H3K27me3) and distal (H3K4me1, H3K27ac, H3K27me3) regions. Each row of heatmap represents one enhancer, with coverage plotted across the 10 kb surrounding enrichment peak. SMARCA4 enrichment at distal regions for forebrain (n = 12,269) (A) and limb (n = 6918) (B). (C) Classified regulatory groups based on histone signatures. (*) Bivalent SMARCA4 elements show a mix of enrichment patterns. (D) Differential enrichment for functional annotation terms associated with distal SMARCA4 regions categorized by histone signature. Shown are the top five enriched “Biological process terms” specific to distal forebrain (blue) and limbs (green) differentially marked SMARCA4 regions. (E,F) Transcription associated with SMARCA4 regions that can be classified as active or repressed based on co-occurring histone marks. RNA-seq data were generated for E11.5 forebrain and limb and the distribution of expression levels of the nearest gene was compared for SMARCA4-enriched regions in forebrain (E) and limb (F) classified by histone signature.











