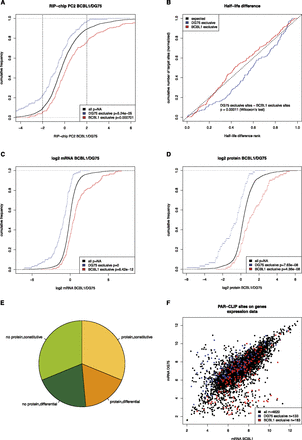
Analysis of context-dependent targets of cellular microRNAs. (A) Distributions of the differential RIP-chip scores as compared to all genes with any PAR-CLIP target sites (see also Fig. 3A). Both targets exclusively active in DG75 as well as in BCBL1 are significantly shifted toward stronger association with RISC in their respective context. The vertical lines indicate a threshold for strongly differentially RISC-associated genes. In both cases, the respective context-dependent targets are more than twofold enriched over the background genes (∼10% of background genes in comparison to >20% of the target genes in both cases). (B) The rank distribution of half-life differences for both sets of context-dependent targets is shown (see also Fig. 3D). BCBL1-specific targets are significantly shifted toward lower half-life difference ranks in comparison to DG75-specific targets indicative for effects of context-dependent microRNA/target interactions in the respective context only. (C,D) The distributions of mRNA and protein fold changes between BCBL1 and DG75 for context-dependent targets of cellular microRNAs, respectively, as compared to the background of all genes with any PAR-CLIP target site. Clearly, based on mRNA as well as on protein levels, context-dependent targets are more highly expressed in their target context. This indicates that the target mRNA expression directly contributes to the cellular context of microRNA-mediated regulation. (E) Depiction of how many genes with context-dependent target sites (n = 311) are constitutively (less than twofold) expressed in both cell lines on an mRNA level and have a protein detected in both cell lines (n = 97), how many are differentially expressed but with a detected protein (n = 54), how many do not have a protein and are differential (n = 63), and how many without a detected protein are constitutive (n = 97). (F) Scatterplot of the microarray intensity measurements for all genes with a PAR-CLIP target site. Interestingly, there seem to be subpopulations of target genes that have extremely low expression values in one of the two contexts and high intensities in the other, where respective target sites are exclusively active. These may indeed correspond to not expressed genes in their inactive target context (see also Supplemental Fig. S4).











