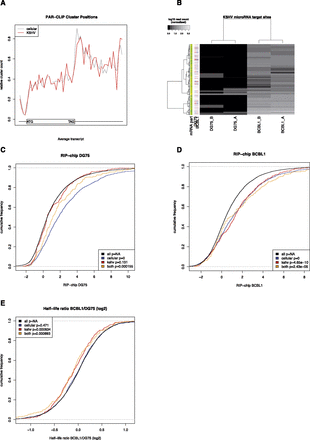
Validation of PAR-CLIP experiments. (A) The distribution of relative positions of target sites on mRNAs is shown. The x-axis represents the average length of 5′ untranslated regions (5′UTR), the coding regions (CDS), and the 3′ untranslated regions of all transcripts with at least one PAR-CLIP cluster. Each transcript was divided into 60 bins, and the relative frequency of target sites falling into each bin is shown on the y-axis. The data clearly illustrate the preferences of target sites in the 3′UTR as compared to CDS and 5′UTR. Viral microRNAs have the same preferences as cellular microRNAs. (B) The normalized number of reads in each cluster (rows) for each of the independent PAR-CLIP experiments (columns) is shown for KSHV microRNA target sites in the four PAR-CLIP libraries. KSHV-negative cell lines (columns 1 and 2) almost exclusively have no reads, whereas for KSHV-positive cell lines, dozens to hundreds of reads are observed per target site. Replicates are highly correlated, indicating high reproducibility. The additional annotations on the left side indicate the part of the transcript where a cluster is located (orange, 5′-UTR; yellow, coding; green, 3′-UTR; gray, not located on known mRNA) and the expression of the transcript in all experiments (red, at least twofold lower expression than the mean expression value for this transcript across all experiments; light red, at least 1.4-fold lower expression than the mean; light blue, at least 1.4-fold higher expression; blue, at least twofold higher expression). We also visualized and inspected individual target sites (Supplemental Fig. S1). (C,D) The log2 RIP-chip enrichment distributions of mRNAs only containing target sites of cellular microRNAs, only containing KSHV microRNA target sites, and containing target sites from both cellular and KSHV microRNAs in the uninfected cell line DG75 and the KSHV positive cell line BCBL1, respectively. KSHV targets are enriched in BCBL1 but not in DG75. (E) The mRNA half-life ratios are shown for the same sets of genes as in C and D. The half-life of mRNAs with KSHV target sites is significantly reduced in BCBL1.











