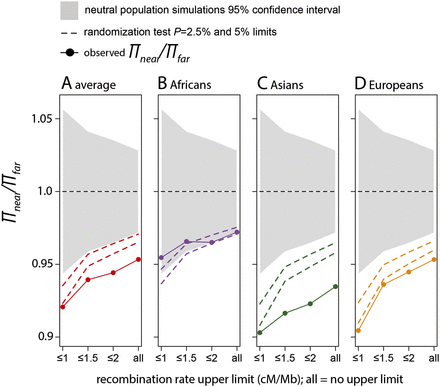
Lower diversity near versus far from amino acid substitutions. Each panel shows the level of synonymous heterozygosity near amino acid changes (πnear) compared with that far from amino acid changes (πfar) for the particular subpopulation. πnear is the average over all near windows (<0.1 cM) from the bootstrap procedure (Methods). πfar is the average over all far windows (>0.5 cM but <1 cM). The gray area depicts the 95% confidence intervals based on neutral simulations (Methods). The dashed lines show 95% and 97.5% confidence intervals based on a randomization test (Methods). Randomization tests result in at most 35% greater variance than the neutral simulations. This is expected given that neutral simulations do not account for complex demography and other sources of noise in the data. On the x-axis, ≤1, ≤1.5, etc., means that we use only windows with recombination rates ≤1 cM/Mb, 1.5 cM/Mb, etc., to compare diversity near and far from amino acid substitutions. (All) All windows are used independently of their recombination rates.











