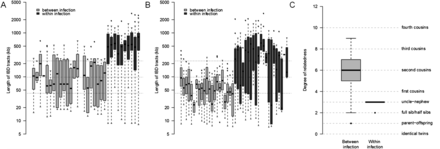
Inferring relatedness within malaria infections. We identified tracts of chromosomes with shared IBD between two parasites for P. vivax (A) and P. falciparum (B) infections using GERMLINE. For P. vivax, between-infection data were generated using both single-cell and bulk sequence from VHX059, VHX333, and WPP1494; and within-host data from WPP1494 alone. For P. falciparum, between-infection data comprises 66 published sequences from western Thailand malaria infections and within-infection data from MKK2664. For clarity, a representative sample of pairwise IBD length distribution is shown in gray with the median and 25th/75th percentiles for the whole population shown by dotted lines. (C) Because we had a sufficiently large population sample for P. falciparum, we used these IBD length distributions to estimate the degree of relatedness between pairs of isolates both between infections and within-infection MKK2664. This showed genomes from within MKK2664 to be significantly more related than population data (χ2 test, χ = 1056.985, P = 8.8 × 10−222).











