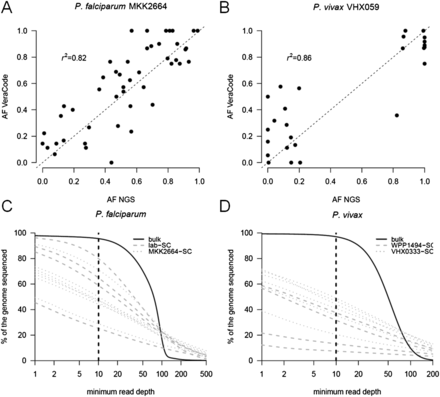
Within-host allele frequency and genome-wide coverage from single-cell sequencing. Using VeraCode genotypes for MKK2664 (A) and VHX059 (B), we estimated the within-host allele frequency of all SNPs that are polymorphic within these respective infections. MKK2664 and VHX059 were also sequenced prior to single-cell sorting and the within-host allele frequencies estimated at each of these loci. We observed a strong correlation (r2 > 0.8) between each of these estimates of within-host allele frequencies in both infections. We estimated the genome-wide coverage we obtained from single-cell sequencing of P. falciparum (C) and P. vivax (D). For each species, the observed coverage profile for a bulk parasite sequence is shown by a solid black line. Full sequencing statistics are given in Supplemental Table 2.











