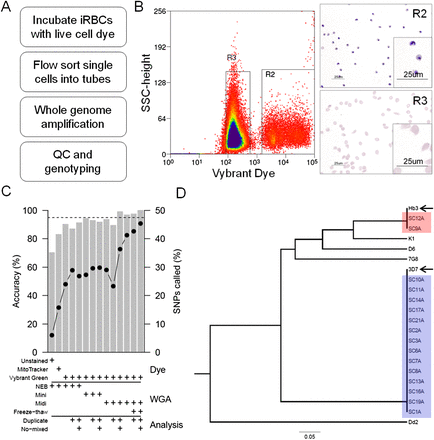
Validation of single-cell genotyping in P. falciparum. (A) The single-cell sorting protocol. (B) We validated that live cell dyes specifically isolate iRBCs from malaria parasite cultures by making giemsa stained smears of cells passing through gates R2 (iRBCs) and R3 (uninfected RBCs)—the full sorting strategy is shown in Supplemental Figure 1. (C) Accuracy (gray bars) and percentage of SNPs called (black dots) of SCG approaches were determined using mixtures of known parasite genotypes. The dyes, WGA kits (PicoPLEX [NEB], REPLI-g Midi and Mini), and analysis parameters are shown below the graph. The rightmost bar shows the optimal protocol determined during our validation. The dashed line denotes 95% accuracy. (D) This optimal protocol accurately clustered single-cell genotypes as 3D7 or Hb3 genotypes without discrepancies. Single-cell genotypes are prefixed “SC” and parental lines named in full and highlighted by arrows. We also include four laboratory strains as genotyping controls: Dd2, 7G8, K1, and D6.











