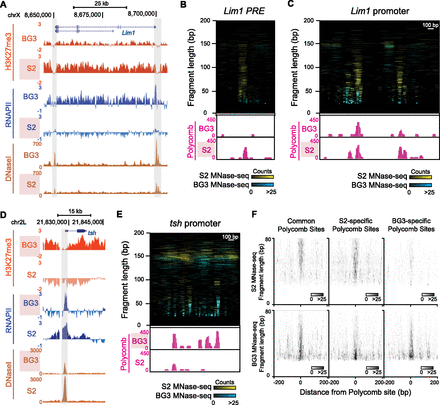
Architectural changes at Polycomb binding sites in alternate repression states. (A) Genome Browser snapshot of a differential H3K27me3 domain that includes the Lim1 gene. The distributions of H3K27me3 (modENCODE X-ChIP), RNAPII (modENCODE X-ChIP), and DNase-seq count density (modENCODE) are shown for S2 cells and BG3 cells. The domain has abundant H3K27me3 methylation and Lim1 is silent in S2 cells, but methylation is reduced and the gene is active in BG3 cells. The two major DHS near Lim1 that correspond to the promoter (right) and a putative PRE (left) are shaded. (B) Comparative plots of MNase-seq fragments (<200 bp) at the Lim1 PRE in S2 (cyan) and BG3 (yellow) cells. Genome Browser snapshots below display normalized read counts from Polycomb N-ChIP (pink) for each cell line. The PRE is extensively occupied in S2 cells, but protection is reduced in BG3 cells. (C) Comparative plots of MNase-seq fragments (<200 bp) at the Lim1 promoter in S2 (cyan) and BG3 (yellow) cells. Browser snapshots below display normalized read counts from Polycomb N-ChIP (pink) for each cell line. The promoter has extensive protection in S2 cells where the gene is repressed. Four Polycomb binding sites are present in S2 cells, only one of which is constitutive and present in BG3 cells. (D) Browser snapshot of a differential H3K27me3 domain that includes the tsh gene, showing the distributions of H3K27me3, RNAPII, and DNase-seq count density for S2 cells and BG3 cells. The domain has H3K27me3 methylation and tsh is silent in BG3 cells, but abolished, and the gene is active in S2 cells. The putative PRE (shaded) defined by DHS and Polycomb binding coincides with the promoter. (E) Comparative plots of MNase-seq fragments (<200 bp) at the tsh PRE in S2 (cyan) and BG3 (yellow) cells. Browser snapshots below display normalized read counts from Polycomb N-ChIP (pink) for each cell line. The PRE is extensively occupied in BG3 cells, but the region is predominantly occupied by nucleosomes in S2 cells. (F) Midpoint plots of MNase-seq fragments at 70 focal Polycomb binding sites common to both S2 and BG3 cells (left), 133 sites found only in S2 cells (middle), and 91 sites found only in BG3 cells (right). Larger fragments are characteristic of Polycomb-bound sites.











