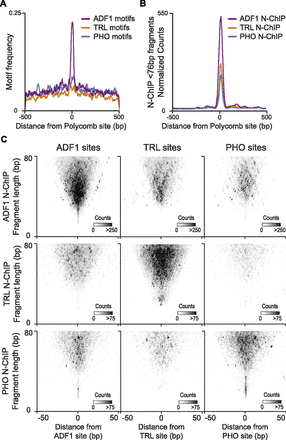
Coincidence of recovered fragments from N-ChIP. (A) Distribution of high-scoring (P < 5 × 10−4) ADF1, TRL, and PHO motifs centered on focal Polycomb binding sites. (B) Distribution of normalized read counts from N-ChIP experiments for ADF1, TRL, and PHO centered on focal Polycomb binding sites. (C) Midpoint plots showing pairwise combinations of ADF1, TRL, or PHO N-ChIP-recovered fragments with their called binding sites. ADF1 sites are predominantly enriched for fragments recovered by ADF1 N-ChIP. TRL sites are enriched for both TRL and ADF1-bound fragments. PHO sites are enriched for both PHO- and ADF1-bound fragments.











