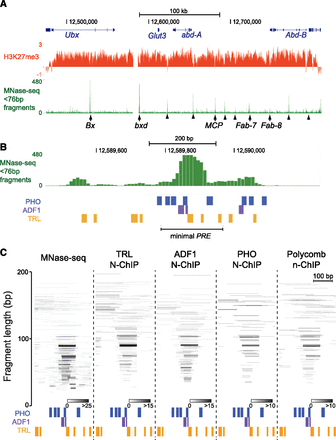
Regulatory elements within the BX-C are clusters of factor-occupied sites. (A) UCSC Genome Browser snapshot of the 330-kb H3K27me3 domain containing the BX-C on chromosome 3R in S2 cells. The distributions of H3K27me3 (modENCODE data) and non-nucleosomal MNase-seq fragments (<76-bp fragments) are shown. Genetically defined PREs (arrows) and regulatory elements inferred from X-ChIP mapping (Schwartz et al. 2006) (arrowheads) are indicated. (B) Browser snapshot of non-nucleosomal MNase-seq fragments at the bxd PRE (chromosome 3R:12,589,450–12,590,250). The positions of high-scoring (P < 5 × 10−4) TRL, PHO, and ADF1 motifs. The segment containing the minimal functional PRE was previously defined (Sipos et al. 2007). (C) Individual fragments in MNase-seq or recovered in the indicated N-ChIP experiments that map to the protected structure (chromosome 3R:12,589,700–12,590,000) at the bxd PRE. Positions of high-scoring (P < 5 × 10−4) TRL, PHO, and ADF1 motifs are indicated. Fragments are positioned on the y-axis by their lengths, and coincident fragments are indicated in shades of gray.











