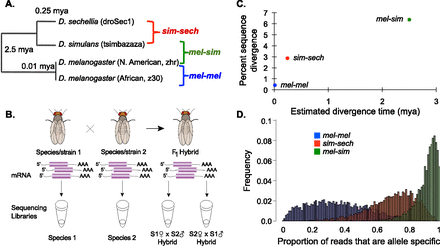
Studying regulatory evolution in the melanogaster group of Drosophila. (A) Phylogenetic relationships and estimated divergence times for the strains and species analyzed are shown. (B) Sequencing libraries for RNA-seq data were derived from mRNA isolated from each species and strain as well as F1 hybrids from reciprocal crosses, in which the maternal and paternal genotypes were reversed (e.g., S1 × S2 and S2 × S1). (C) The percent sequence divergence observed in the regions of the genome used to map RNA-seq reads (y-axis) is compared with published estimates of divergence time (x-axis). (D) The proportion of reads from each gene that is allele-specific is shown for the mel-mel (blue), sim-sech (red), and mel-sim (green) comparisons.











