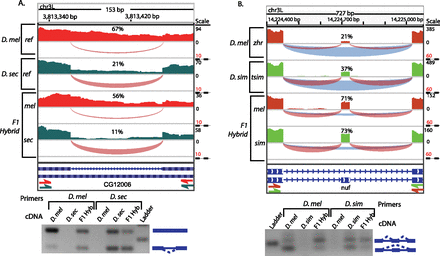
Cis- and trans-acting differences in alternative splicing. IGV Genome Browser images show RNA-seq read coverage, depicted in a bar graph (above). Arcs depict splice-junction read coverage (per million mapped junction reads). The width of each arc represents the relative number of junction reads. For exon skipping, exclusion junctions are shown in blue and inclusion junctions are shown in red. The percent spliced in (PSI) for each sample is shown above the alternative event. Gel images show the results of RT-PCR experiments using species-specific primers. (A) An intron in CG12006 is retained much more frequently in D. melanogaster than in D. sechellia. In F1 hybrids, allele-specific splicing differences show that the divergence is predominantly cis-acting. (B) An exon in nuf is skipped more frequently in D. melanogaster than in D. simulans. Allele-specific RNA-seq from F1 hybrids shows equal inclusion levels, suggesting that divergence in nuf splicing is predominantly trans-acting.











