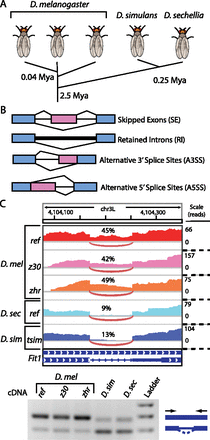
Comparing splicing regulation among Drosophila species. (A) Comparison of divergence times for strains and species used. (B) The four major classes of alternative splicing. Constitutive and alternative exonic regions are depicted in blue and pink, respectively. (C) Example of cross-species comparison of intron retention using RNA-seq. (Top) IGV Genome Browser tracks depict differences in read coverage within the retained intron in each strain and species. The Fit1 intron is more frequently retained in all strains of D. melanogaster, compared with D. simulans and D. sechellia. (Bottom) RT-PCR using primers spanning the retained intron correlates with RNA-seq estimates of divergent splicing.











