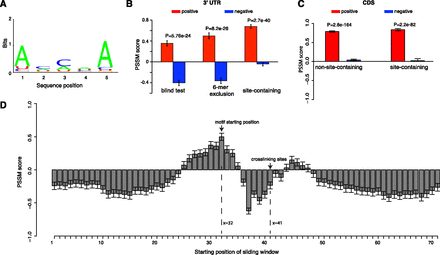
AGO recognizes its targets in a sequence-specific manner within the highly accessible region upstream of the crosslinking site. (A) The mRNA fragments that are depleted of miRNA binding sites have an overrepresented sequence motif within the ∼10-nt accessible regions. (B) Discriminative power of the motif among three independent test sets. (Left) A subset of the AGO-bound mRNAs that was not used in deriving the motif; note these mRNAs were depleted of miRNA binding sites. (Middle) The same data set after removing sequences with at least one 6-mer match to any miRNA seed region. (Right) AGO-bound mRNA sequences that are highly enriched for miRNA binding sites. Overrepresentation of the motif in each data set was assessed by the sum of the position-specific scoring matrix (PSSM) scores for its letters. The more positive scores indicate a greater enrichment for the sequences resembling the motif, while the negative scores indicate the depletion of this motif. (C) Discriminative power of the motif among the AGO-bound sequences in coding regions with (site-containing) and without (non-site-containing) downstream miRNA binding sites. (D) Nine-nucleotide sliding window scan for the AGO-bound sequences that were not used to derive the AGO motif. These sequences were centered at the crosslinking site at position 41. The x-axis is the starting position of the sliding window, and the y-axis is the PSSM score for the AGO motif inside each sliding window. The motif is strongly favored in the ∼10 nt immediate upstream (x = 32) of the crosslinking sites (x = 41) and is disfavored in most other regions (receiving negative PSSM scores). Error bars, 1 SEM.











