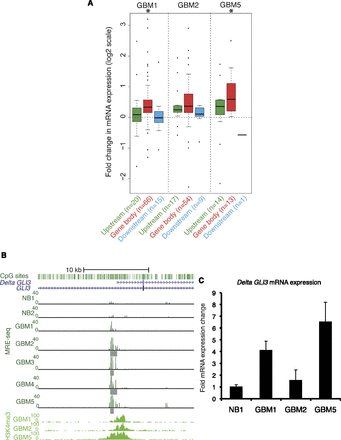
A subset of recurrent DNA hypomethylation coincides with H3K4me3 in the same primary tumor. (A) Gene expression change for genes nearest each DNA hypo/K4me3 locus, which, taking into account the direction of transcription, can be located upstream (5′), within the gene body, or downstream (3′). Hypomethylated DMRs at Q < 10−5 and H3K4me3 peaks (P < 1 × 10−10) were used to define DNA hypo/K4me3 loci. The Affymetrix GeneChip Human Gene 1.0 ST expression changes (GBM-NB) for the nearest RefSeq gene are shown as box plots on a log2 scale. The number of genes analyzed in each box plot is given along with the labels at the bottom. Statistically significant gene expression changes were determined using a t-test at P < 0.05 (asterisks above box plots). A total of 19,628 Entrez genes were analyzed on the array. (B) Recurring colocalization of DNA hypomethylation and H3K4me3 at the Delta GLI3 transcript promoter. Individual hypomethylated DMRs are shown under the MRE. (C) Quantification of Delta GLI3 transcript abundance by isoform-specific qRT-PCR. Fold expression ± SD relative to normal brain is graphed for a representative experiment. (NB) Normal brain.











