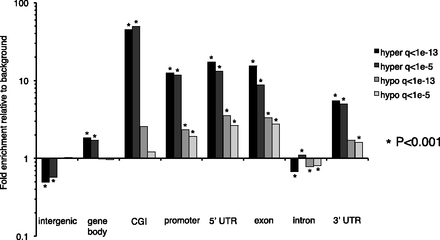
Enrichment for genomic features among recurring hyper- and hypomethylated DMRs at Q < 10−13 and Q < 10−5. Enrichment was calculated using the background distribution of CpG-containing 500-bp windows on autosomes that were also used for M&M analysis. Statistically significant enrichment (or depletion) was calculated using a binomial test, and those significant at P < 0.001 are indicated with asterisks. The definitions of genomic features are the following: intergenic: regions between 3′ transcription end site (TES) of a gene to the 5′-most TSS of next gene; gene body: 5′-most TSS of a gene to 3′-most TES of the same gene; CGIs: from UCSC annotation; promoter: −2.5 to +0.5 kb from the 5′-most TSS. 5′ UTRs, exons, introns, and 3′ UTRs were defined from the RefSeq database. Note that some of these genomic features are not mutually exclusive.











