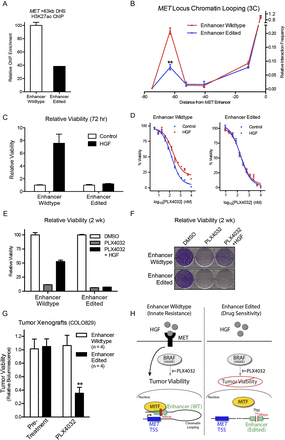
Targeted MET enhancer disruption suppresses innate resistance to inhibition of the BRAF oncokinase. (A) H3K27ac ChIP-qPCR at the MET +63-kb enhancer of wild-type (WT) versus enhancer-edited (EE) cells. (B) 3C of WT versus EE cells at the MET locus. (**) P = 1.00 × 10−3. (C) Viability quantification at 72 h of D, WT and EE cells in PLX4032 dose-response curve ± exogenous HGF. (E) Viability quantification at 2 wk in vitro of WT and EE cells in PLX4032 ± exogenous HGF. (F) Cresyl violet staining of cell populations in E. (G) Tumor cell viability in vivo assessed by bioluminescent signal in COLO829 tumor xenografts before and after treatment with BRAF inhibitor PLX4032. (**) P = 3.49 × 10−3; n = 4 mice/group. (H) Hypothetical working model for MITF-dependent induction of HGF/MET-mediated innate resistance to BRAF (left), and the disruption of this process by targeted genomic editing of the MET +63-kb enhancer (right).











