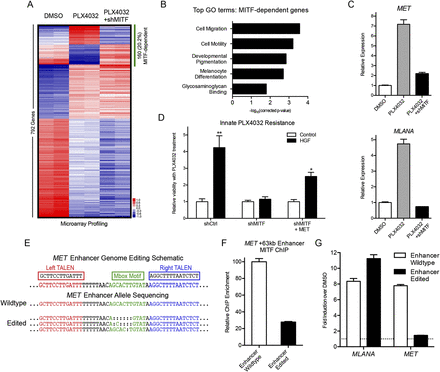
Enhancer genome editing decouples BRAF inhibitor-driven induction of MET and melanoma differentiation. (A) Mean-centered heatmap of significantly changed genes in COLO829 melanoma cells treated with vehicle control (DMSO), PLX4032 BRAF inhibitor, and PLX4032 plus shRNA to MITF. MITF-dependent gene expression changes are highlighted by the green bar. (B) Top Gene Ontology (GO) terms from MITF-dependent genes in A. (C) Expression of MET (top) and the melanocyte differentiation antigen MLANA (bottom) in COLO829 melanoma cells as a function of PLX4032 and MITF depletion. (D) Viability of COLO829 melanoma cells in response to PLX4032 in the presence or absence of exogenous HGF, MITF depletion, and MET overexpression. (**) P = 0.0043; (*) P = 0.0152. (E) Schematic of the MET +63-kb enhancer TALEN targeting strategy; the Mbox sequence motif is shown in green. The wild-type (WT) normal allele found in COLO829 and the enhancer-edited (EE) clone alleles are shown. Deep sequencing validated that no WT alleles remained in the EE clone. (F) MITF ChIP-qPCR at the MET +63-kb enhancer of WT versus EE cells. (G) Induction of MLANA and MET expression by PLX4032 in WT and EE cells.











