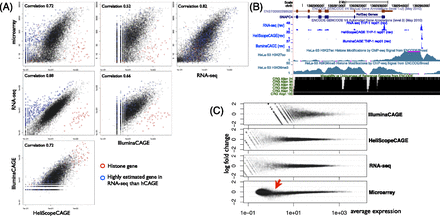
Gene expression quantification by using different platforms. (A) Scatter plot of gene expressions by using different platforms, where RPKM (reads per kilobase of exon models per million) is used for RNA-seq. The Spearman’s correlation coefficient is shown for individual comparisons. (B) Individual profiles in SNAPC4 locus. Blue signals indicate reverse-strand signals by the CAGE and RNA-seq platforms. ENCODE histone modification profiles indicating promoter and elongation activities are shown below. (C) MA plot between the 50% mixture experimental profile and the computationally synthesized one from the THP-1 and HeLa profiles. The 50% profile is based on the average of triplicates, while the computational one is based on combining six profiles (triplicates of THP-1 and HeLa).











