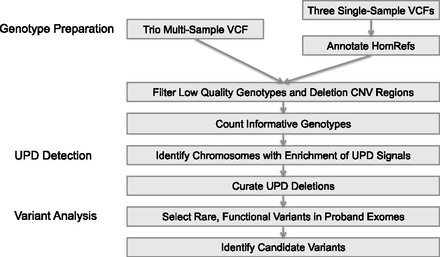
Study workflow. The study consisted of three main steps: data preparation, UPD detection, and candidate variant analysis. In the data preparation stage, we collected informative genotypes seen in all members of each trio. Either a multi-sample trio VCF or three single-sample VCFs can be used as input; the latter requires the annotation of homozygous reference genotypes, not usually encoded in single-sample VCF files. In the UPD detection stage, we selected trios containing a proband chromosome with an enrichment of UPD-informative genotypes. Exomes (five) available for samples with a detected UPD event were selected for the candidate workup analysis, in which we attempted to find rare protein-altering variants that may manifest in the proband’s phenotypes.











