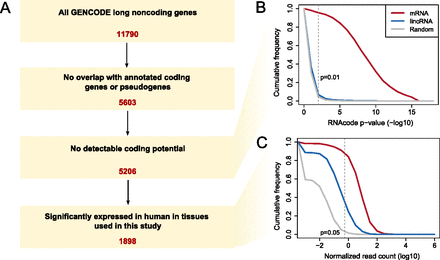
Definition of the lincRNA set. (A) Filtering steps of all GENCODE noncoding transcripts to the final set of lincRNAs used for further analysis in this study. (B) Cumulative distribution of RNAcode (Washietl et al. 2011) P-values measuring the coding potential of transcripts. The P-value cutoff of 0.01 is indicated, and for comparison the distributions for coding transcripts and randomized transcripts are also shown. (C) Distribution of normalized expression levels in human. The maximum FPKM (fragments per million reads per kb of transcript) over all tissues is shown. The cutoff was chosen empirically using randomized transcripts (Methods) as the background distribution and requiring a significance level of 0.05. If read counts were zero, we set the count to 10−3, explaining the discontinuous shape of the curves.











