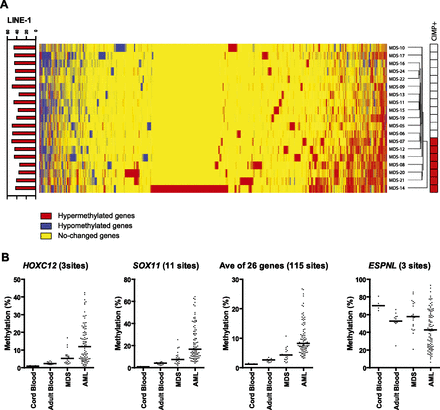
DNA methylation analyses in humans. (A) Hierarchical clustering analysis of human MCAM. Heat-map analysis showing the MCAM results in 19 MDS patients. Red, yellow, and blue correspond to hypermethylated, nonchanged, and hypomethylated loci, respectively. All probes that were unchanged in all samples were excluded from the analysis. The bar graph on the left shows the methylation percentage of LINE-1 by pyrosequencing analysis. Red-colored boxes on the right side represent the CIMP-positive case defined by MCAM assay. (B) DNA methylation profiles by DREAM analysis in human cord blood, adult blood, MDS, and AML patients. The percentages of methylated cytosines in the samples as obtained from DREAM analysis. Each dot corresponds to one individual. We show here data on HOXC12, SOX11, and ESPNL. The averaged data were derived from the methylation data of 26 hypermethylated genes listed in Supplemental Table S14. The bar in the graphs represents the median.











