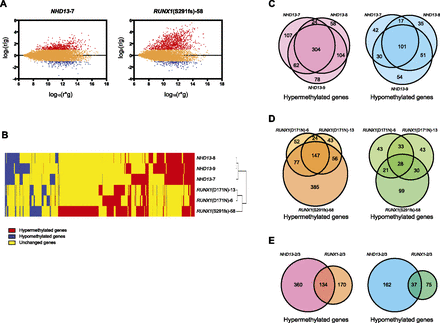
MCAM analysis in mouse MDS models. (A) R-I plot of the probes with FDR at 5% and a fold change greater than two for MCAM. An R-I plot displays the log2(R/G) ratio for each element on the array as a function of the log10(R × G) product intensities and can reveal systematic intensity-dependent effects in the measured log2 (ratio) values. The red and blue spots indicate probes hypermethylated and hypomethylated in diseased samples, respectively. (B) Hierarchical clustering analysis of mouse MCAM. Heat-map analysis showing the MCAM results in six model mice. Red, yellow, and blue correspond to hypermethylated, nonchanged, and hypomethylated loci, respectively. For clarity we excluded probes that were unchanged in all samples. (C) Venn diagram of individual differences of hyper- and hypomethylated genes in NHD13 mice analyzed by MCAM. Each circle represents one individual. (D) Venn diagram of mutational differences of hyper- and hypomethylated genes in RUNX1 mice analyzed by MCAM. Each circle represents one individual. (E) Venn diagram of hyper- and hypomethylated genes overlapped between NHD13 and RUNX1 mice analyzed by MCAM. Each circle represents a genotype.











