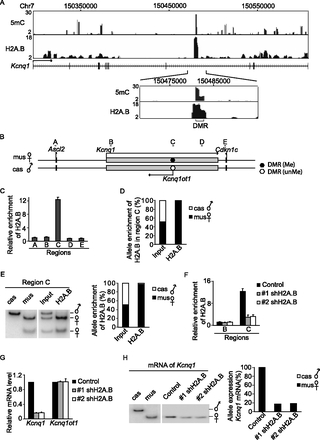
H2A.B regulates the transcription of the imprinted Kcnq1 locus. (A) ChIP-seq and MeDIP-seq profile of H2A.B and 5mC in Kcnq1 loci. The x-axes indicate the analyzed genomic region. The y-axes represent the fold enrichment of 5mC or H2A.B compared to the irrelevant IgG control. Enlarged view shows the colocalization of H2A.B and 5mC in the DMR region. (B) Schematic sketch of the Kcnq1 locus. (C) ChIP analysis confirms that H2A.B is incorporated into the DMR of the Kcnq1 locus but not other regions. Data are presented as mean ± SEM (n = 4). (D,E) H2A.B is enriched in the maternal DMR of Kcnq1. ChIP assays are performed using anti-H2A.B antibody. Based on the SNPs between M. m. musculus (mus) and M. m. castaneus (cas), allele-specific deposition of H2A.B is determined by DNA sequencing (D) or the BmgBI digestion of an amplified DNA fragment into different patterns (E). (F) shRNA treatment depletes H2A.B at the DMR of Kcnq1. ChIP analyses of H2A.B at the DMR and the adjacent region are performed. Data are presented as mean ± SEM (n = 4). (G) Knockdown of H2A.B suppresses the transcription of the Kcnq1 gene. Data are presented as mean ± SEM (n = 4). (H) The transcription of Kcnq1 is dependent on the maternal allele and H2A.B. Allele-specific expression of imprinted Kcnq1 was determined by different NlaIII digestion patterns based on the SNP between mus and cas.











