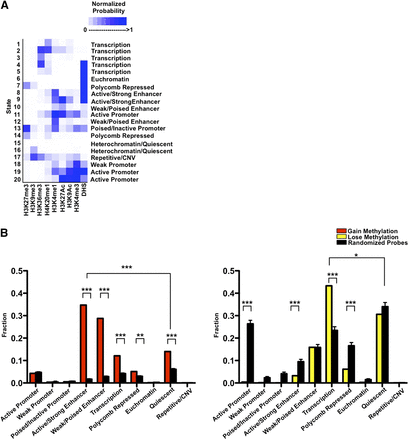
Alteration of the epigenetic landscape in the neobladder occurs predominantly in the enhancers and transcribed regions. (A) Chromatin states for the normal small intestine are defined based on a 20-states chromHMM model using publicly available data, and underlying chromatin states were determined for each CpG probe. Color scale measures the normalized probability of finding a specific histone mark in a particular chromatin state, and each state is annotated based on the levels of enrichment of each histone mark. (B) Chromatin states were collapsed into 10 distinct states as plotted on the x-axis. A cluster of 4162 probes that gain methylation (left panel) and a cluster of 748 probes that lose methylation (right panel) were compared to randomized sets of an equal number of loci where 1000 trials were performed, and the P-value was determined using a binomial test. ([***] P < 2.2 × 10−16, [**] P < 3.3 × 10−13). The enhancer states are significantly enriched in the cluster of probes that gains methylation, whereas the cluster of probes that loses methylation predominantly falls in the transcribed regions.











