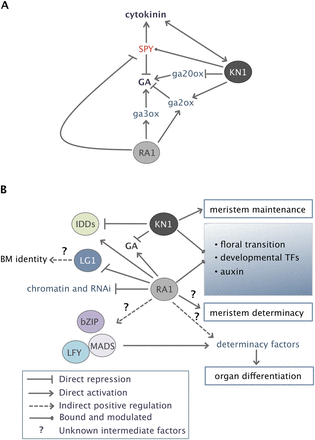
Models for RA1-mediated regulation and integration with KN1-based meristem maintenance pathways. (A) RA1 and KN1 interact via gibberellic acid (GA) biosynthesis and signaling. RA1 may modulate GA levels in a spatiotemporal manner by activating genes for its biosynthesis and catabolism and negatively regulating a repressor of GA signaling, SPY. (B) RA1 interfaces with various developmental and regulatory networks, and interacts with KN1-based meristem maintenance via common targets and pathways. RA1 directly represses genes involved in chromatin and RNAi and positively regulates a suite of co-expressed determinacy factors. Promoters of the latter were enriched for binding sites of LFY, bZIP, and MADS-box TFs, and therefore activation of determinacy factors by RA1 could work in part through coregulation by these TFs. RA1 positively regulates a set of IDD TFs, including one that is co-bound and repressed by KN1, and negatively regulates lg1, which may play a role in BM identity, possibly by establishing a boundary. RA1 and KN1 also co-target genes related to floral transition, auxin biology, and the integration of environmental and developmental cues.











