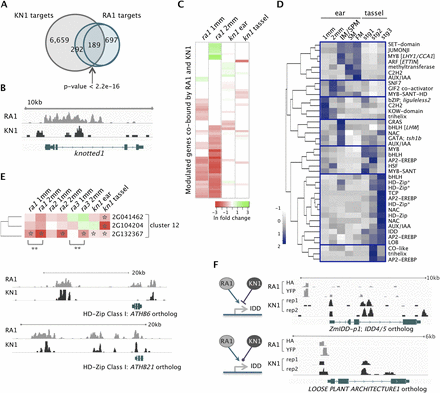
Integration of RA1- and KN1-dependent networks. (A) RA1 and KN1 bound 481 shared target genes (189 at the same genomic position), which was greater than expected by chance based on Fisher's test. (B) RA1 bound kn1 in its third regulatory intron. (C) Sixty-five targets were cobound by RA1 and KN1 at the same genomic position and differentially expressed in ra1 and/or kn1 loss-of-function mutants; these genes tended to have stronger dependence on RA1 than KN1 for their normal expression; (green to red) up- to down-regulation; (ln) natural log. (D) Expression profiles for 40 TFs cobound by RA1 and KN1 at overlapping genomic regions showed signatures of spatiotemporal regulation. TFs are listed by their family or protein domain name and, where provided, Arabidopsis ortholog name in brackets. (E) Three co-expressed HD-Zip Class I genes (indicated by an asterisk in D) were modulated targets of RA1 and/or KN1. All were significantly down-regulated ([*] P < 0.05) in kn1 tassels; GRMZM2G132367 was significantly down-regulated in ra1, ra2, and ra3 mutant ears by 2 mm and showed significant change ([**] P < 0.001) between 1 and 2 mm in ra1 and ra3, but its expression remained unchanged in wild-type ears from 1 to 2 mm. RA1 and KN1 also cobound putative intergenic regulatory regions ∼15 kb upstream of these HD-Zip genes. Shown are orthologs of ATHB6 (GRMZM2G132367) and ATHB21 (GRMZM2G104204). (F) IDD genes bound by both RA1 and KN1 were positively modulated by RA1. ZmIDD-p1 (GRMZM2G179677) was repressed by KN1 while expression of the LOOSE PLANT ARCHITECTURE 1 ortholog (GRMZM2G074032) was not significantly altered in kn1 mutants.











