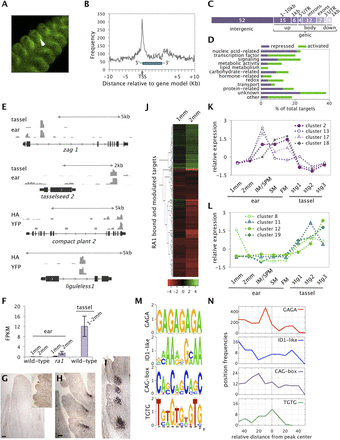
Genome-wide binding profiles for RAMOSA1. (A) YFP-tagged RA1 is expressed in an adaxial domain subtending SPMs in developing inflorescences, and localized to the nucleus. (B) Distribution of RA1 binding relative to maize gene models showed strong enrichment −1.5 and +1.5 kb from the TSS. (C) Distribution of high-confidence peak summits across genomic features (numbers are based on percent of total). Within a genic region, (up) upstream; (body) gene body; (down) downstream. (D) Bound and modulated targets of RA1 grouped by functional class. (E) RA1 bound genes with known inflorescence phenotypes; zag1, ts2, ct2, and lg1. Examples of overlapping peaks in ear and tassel (zag1 and ts2) and HA- and YFP-tagged libraries (ct2 and lg1) are shown. (F) lg1 was up-regulated in ra1 mutant ears and in wild-type tassel primordia compared with wild-type ears. (G) Immunolocalization of the LG1 protein indicates its absence in wild-type ears (inset image shows determinate SPMs). (H) In ra1 mutant ears, LG1 is localized to the adaxial side of developing branch meristems and (I) in wild-type tassels is localized to the base of long branches. (J) Bound and modulated targets of RA1 were more strongly regulated at 2 mm compared with 1 mm. (K,L) Expression profiles represent cluster centers from Figure 3A: Repressed targets were largely co-expressed in clusters 2, 13, 17, and 18; activated targets were associated with clusters 8, 11, 12, and 19. (M) Analysis of high-confidence RA1 binding sites within gene promoters showed enrichment of de novo motifs: a GAGA-repeat element, a motif similar to the indeterminate1 (id1) binding site (P = 5.6 × 10−9), novel CAG-box and TG repeat motifs. The latter two were most strongly enriched in promoters of activated genes. (N) Motifs were enriched at specific positions relative to the center of RA1 binding sites.











