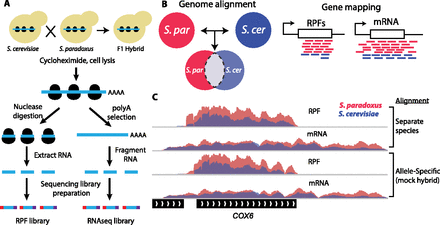
Overview of allele-specific ribosome profiling to measure divergence in ribosome occupancy, mRNA abundance, and translational efficiency. (A) Ribosome profiling was performed on log-phase cultures of S. cerevisiae, S. paradoxus, and their F1 hybrid. Ribosome protected fragments (RPFs) were purified and cloned into Illumina high-throughput sequencing libraries (left). Poly-adenylated mRNA sequencing libraries were prepared in parallel (right). (B) Sequence reads from each sample were aligned to both species genomes. Allele-specific reads were identified by comparing genomic alignments and mapped to corresponding regions of orthologous ORFs. (C) Comparison of separate and allele-specific coverage in ribosome profiling experiments. IGV browser tracks showing normalized coverage of RPF and mRNA sequence reads from S. cerevisiae (blue) and S. paradoxus (magenta) over the COX6 gene (YHR051W). Measurements of interspecies differences in read coverage are equivalent for separate species (upper) and allele-specific alignments (“mock hybrid,” lower) (see also Supplemental Fig. S1).











